
Revista UNICIENCIA
Uniciencia Vol. 38(1), January-December, 2024
E-ISSN: 2215-3470
DOI: https://dx.doi.org/10.15359/ru.38-1.18
A Molecular Docking and Dynamics Simulation Study on Prevention of Merozoite Red Blood Cell Invasion by Targeting Plasmodium vivax Duffy Binding Protein with Zingiberaceae Bioactive Compounds
Estudio de simulación dinámica y de acoplamiento molecular sobre la prevención de la invasión de glóbulos rojos de merozoitos atacando la proteína de unión al antígeno Duffy de Plasmodium vivax con compuestos bioactivos de Zingiberáceas
Estudo de simulação dinâmica e de acoplamento molecular na prevenção da invasão de glóbulos vermelhos por merozoítos, visando a proteína de ligação Duffy do Plasmodium vivax com compostos bioativos de Zingiberaceae
Muhammad Fikri Heikal1, Wira Eka Putra2*, Sustiprijatno3, Arief Hidayatullah4, Diana Widiastuti5, Mardiana Lelitawati1
Received: Aug/30/2023 • Accepted: Feb/7/2024 • Published: Jul/31/2024
|
Abstract [Objective] Plasmodium vivax predominantly infects many people in numerous tropical areas, including Southeast Asia, the Western Pacific, the Americas, and the Eastern Mediterranean. The uniqueness of forming dormant stages can lead to relapse in vivax malaria upon further infection. This study used molecular docking and dynamic simulation to predict potential bioactive compounds from the Zingiberaceae plant family as inhibitors by targeting Plasmodium vivax Duffy Binding Protein (PvDBP). PvDBP-DARC molecular interaction is required to mediate the merozoite invasion process into red blood cells. Inhibiting this process can possibly control parasite growth and development. [Methodology] Molecular docking screening was conducted by using 138 natural compounds from the Zingiberaceae plant family targeting Plasmodium vivax Duffy binding protein (PvDBP). The top two compounds with the lowest binding energy were selected to be analyzed by pharmacokinetics prediction and molecular dynamic (MD) simulation. [Results] Molecular docking screening resulted in the top two compounds with the lowest binding energy value, including 5,7-dihydroxyflavanone (-9.3 kcal/mol) and pinostrobin (-9.2 kcal/mol). These compounds are predicted to have stronger interaction than chloroquine as a control. Furthermore, the potential compounds also interact with DARC binding site residues and maintain them during the molecular dynamic simulation process. Otherwise, chloroquine as a control cannot retain 75% binding residues towards PvDBP. A molecular dynamic study revealed that all three complexes have relatively similar stability. [Conclusions] We predicted that the two bioactive compounds (5,7-dihydroxyflavanone and pinostrobin) have the potential as merozoite invasion inhibitors. Keywords: anti-malaria drug, in silico, Plasmodium vivax, PvDBP, Zingiberaceae. Resumen [Objetivo] Plasmodium vivax infecta ampliamente a muchas personas en algunas regiones. La singularidad de formar una etapa latente hace que la malaria vivax pueda ser inducida por una recaída por una infección adicional. En el presente estudio, utilizamos un enfoque de simulación dinámica y acoplamiento molecular para predecir la posibilidad de que compuestos bioactivos de la familia de plantas Zingiberaceae sean candidatos a fármaco contra la malaria dirigiéndose a la proteína de unión a Duffy de Plasmodium vivax (PvDBP). Se requiere la interacción molecular PvDBP-DARC para mediar el proceso de invasión de merozoitos en los glóbulos rojos. La inhibición de este proceso posiblemente pueda controlar el crecimiento y desarrollo del parásito. [Metodología] El análisis de acoplamiento molecular dio como resultado los dos compuestos principales con el valor de energía de unión más bajo, incluida la 5,7-dihidroxiflavanona (-9,3 kcal/mol) y la pinostrobina (-9,2 kcal/mol). [Resultados] Estos compuestos previstos tienen una interacción más fuerte que la cloroquina como control. Además, los compuestos potenciales también interactúan con los residuos del sitio de unión de DARC y lo mantienen durante el proceso de simulación de dinámica molecular. De lo contrario, la cloroquina como control no puede retener el 75% de los residuos de unión hacia PvDBP. El estudio de dinámica molecular reveló que los tres complejos tienen una estabilidad relativamente similar. [Conclusiones] Predijimos que los dos compuestos bioactivos tienen potencial como inhibidores de la invasión de merozoitos. Palabras clave: Medicamento contra la malaria; en silicio; Plasmodium vivax; PvDBP; Zingiberáceas. Resumo [Objetivo] O Plasmodium vivax infecta amplamente muitas pessoas em algumas regiões. A singularidade de formar um estágio latente significa que a malária vivax pode ser induzida por recaída devido à infecção adicional. No presente estudo, utilizamos uma abordagem de simulação dinâmica e de acoplamento molecular para prever a possibilidade de compostos bioativos da família de plantas Zingiberaceae como candidatos a medicamentos antimaláricos, visando a proteína de ligação Duffy do Plasmodium vivax (PvDBP). A interação molecular PvDBP-DARC é necessária para mediar o processo de invasão de merozoítos nos glóbulos vermelhos. A inibição deste processo pode possivelmente controlar o crescimento e desenvolvimento do parasita. [Metodologia] A análise de acoplamento molecular resultou nos dois principais compostos com o menor valor de energia de ligação, incluindo 5,7-dihidroxiflavanona (-9,3 kcal/mol) e pinostrobina (-9,2 kcal/mol). [Resultados] Esses compostos previstos têm uma interação mais forte do que a cloroquina como controle. Além disso, os compostos potenciais também interagem e mantêm resíduos do sítio de ligação do DARC durante o processo de simulação de dinâmica molecular. Caso contrário, a cloroquina como controle não pode reter 75% dos resíduos de ligação ao PvDBP. O estudo de dinâmica molecular revelou que os três complexos apresentam estabilidade relativamente semelhante. [Conclusões] Previmos que os dois compostos bioativos têm potencial como inibidores da invasão de merozoítos. Palavras chave: Medicamentos antimalária; em silício; Plasmodium vivax; PvDBP; Zingiberáceas. |
Plasmodium vivax is a virulent malaria parasite that widely spreads in Southeast Asia, the Americas, the Eastern Mediterranean, and the Western Pacific (Howes et al., 2016; World Health Organization, 2020). In 2019, this parasite became the dominant species that caused malaria disease in Southeast Asian regions, accounting for more than 50% of cases, which totaled 6.3 million (World Health Organization 2020). Furthermore, 80% of the 1.6 billion population in Southeast Asia has a risk of vivax malaria transmission (Battle & Baird, 2021). Unlike Plasmodium falciparum, this species has a unique dormant stage by forming hypnozoites. A hypnozoite is a dormant form of parasite that is activated after further infection (Dini et al., 2020; Markus, 2011). This uniqueness causes vivax malaria to be called a relapsing malaria disease.
In malaria, blood infection becomes a crucial stage to expand parasite invasion and increase its population. Inhibition at this stage probably prevents important life cycle continuation, such as gametocyte, trophozoite, and schizogony stage development. The success of Plasmodium vivax in invading red blood cells depends on the existence of a Duffy-binding antigen protein (Howes et al., 2016). Hence, host-pathogen molecular interaction through the Duffy antigen protein during the blood stage is required to support the parasite invasion process. One of the parasite’s proteins included in the mechanism is the Plasmodium vivax Duffy Binding Protein (PvDBP). This protein is a member of the erythrocyte binding–like (EBL) protein superfamily, consisting of 330 amino acids and twelve conserved cysteine residues (Batchelor et al., 2011; Hans et al., 2005). The PvDBP is localized in the micronemes organelle and released during the invasion process (Han et al., 2016). The PvDBP binds to Duffy antigen receptor chemokines (DARC) driven via the dimerization process (Batchelor et al., 2011). This interaction mediates merozoite internalization into red blood cells. Because of its vital role in parasite development, inhibition of PvDBP-DARC interaction may greatly contribute to parasite control.
Malaria treatment using chloroquine is still the mainstay, especially for vivax malaria in several regions (Chu & White, 2021; Ould et al., 2015; Sutanto et al., 2010). Even though multiple studies revealed that chloroquine reduces its efficacy to cure vivax malaria disease due to resistance occurrence (Ferreira et al., 2021; Getachew et al., 2015; Htun et al., 2017), other studies reported that Plasmodium vivax also resists several antimalaria drugs, including pyrimethamine and sulfadoxine (Korsinczky et al., 2004; Ratcliff et al., 2007). Therefore, the emergence of Plasmodium vivax resistance becomes a challenge in identifying novel therapeutic agents, especially from natural resources.
The study of plant metabolites to find out their potential as drug candidates for many diseases has been encouraged. Several reports demonstrated that secondary metabolites from plants show positive results in decreasing malaria parasite infection (Abdou et al., 2022; Ahmed et al., 2022; Okokon et al., 2022). The Zingiberaceae plant family is one of the popular plants widely used in traditional medicine to cure some diseases. Almost all parts of this plant potentially contain beneficial bioactive compounds. Therefore, the evaluation of its potential as a drug candidate, especially for vivax malaria, must be expanded.
In this study, we use a bioinformatics approach to predict interactions between bioactive compounds from the Zingiberaceae plant family targeting the PvDBP. We generally use two types of analysis, including molecular docking and dynamic simulation. We analyze compounds-protein interaction types and strength by using molecular docking, whereas molecular dynamic simulation is used to analyze the protein behavior of PvDBP-ligand complexes during simulation time.
● Protein Target Retrieval
A PvDBP 3D structure file (PDB ID: 6R2S) in PDB format was obtained from RSCB PDB database (https://www.rscb.org/). Before entering the docking process, we removed all native ligands that bind into protein using PyMOL v.2.5.2 software (https://www.pymol.org).
● Ligand Selection and Preparation
Approximately 200 bioactive compound data were collected from 15 Zingiberaceae plant species, as in our previous study (Heikal et al., 2023). The data were referred from several previous reports. Moreover, bioactive compound SDF files were downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov), in accordance with our previous studies (Hidayatullah et al., 2022; Hidayatullah et al., 2021; Putra, 2018; Putra et al., 2017). To examine compounds’ physical and chemical properties, we performed a drug-like test using the Lipinski rule of five web server (http://www.scfbio-iitd.res.in/software/drugdesign/lipinski.jsp). There are about five indicators used as key factors in determining the drug-likeness of ligands, including hydrogen bond donor, hydrogen bond acceptor, molecular weight, lipophilicity, and molar reactivity (Lipinski, 2004).
● Molecular Docking Screening
Molecular docking procedures were followed in our previous studies (Widiastuti et al., 2023; Hidayatullah et al., 2021; Putra & Rifa’I, 2020; Putra et al., 2020). The PyRx integrated with autodock vina (https://pyrx.sourceforge.io/) was used for the docking screening process. After 137 compounds passed drug-like testing, they were minimized and converted into PDBQT format. Chloroquine as vivax malaria was used as control. The grid size used was X: 66.2301, Y:-104.93, Z:-9.2953, and the coordinate dimension (Angstrom) was X: 84.4322, Y: 58.3046, Z: 57.4419.
● Data Visualization
We performed two types of visualization processes (Putra et al., 2023; Hidayatullah et al., 2021; Putra et al., 2019). The top two compounds + control – PvDBP complexes were visualized to determine the type of interaction and binding region. We used LigPlot+ v.2.2 (https://www.ebi.ac.uk/thornton-srv/software/LigPlus) for 2D visualization and PyMOL v.2.5.2 (https://www.pymol.org) for 3D visualization.
● Molecular Dynamics Simulation
We performed a molecular dynamic simulation of three PvDBP-ligand complexes in 1000 picoseconds simulation time similar to our previous simulation setting (Maslikah & Putra, 2024; Hidayatullah et al., 2023; Sari et al., 2023; Ashar et al., 2023). The complexes were prepared according to the physiological state (pH 7.4, 0.9% salt condition, 37 oC, and 1 atm). We used MD macro programs for the MD simulation running process and MD analysis, along with MD macro program analyzers, for the molecular dynamics data analysis.
● Pharmacokinetics Prediction
To predict the adsorption, distribution, metabolism, excretion, and toxicity (ADMET) properties of the selected compounds, we performed pharmacokinetics prediction using the pkCSM webserver (https://biosig.lab.uq.edu.au/pkcsm/). The prediction of the pharmacokinetics parameters is important in drug discovery due to its contribution to initial results that could be referred for further drug development.
About 170 amino acids were identified as DARC binding site residues of PvDBP. All three compounds partially bind to the crucial binding residue (Table 1). 5.7-Dihyroxyflavanone (CID: 238782) has the lowest binding energy value (-9.3 kcal/mol) and pinostrobin (CID: 6950539) is the second (-9.2 kcal/mol). Around 80% of both binding residues are DARC binding residues. In contrast, chloroquine as control has a higher binding energy value (-5.5 kcal/mol) with 50% residues are DARC binding residues. We predict that the attachment of two potential compounds and control can disturb PvDBP-DARC molecular interaction during merozoite invasion (Figure 1).
Table 1. Binding residue details. Bold indicates Duffy Antigen Membran Chemokines (DARC) binding residue.
|
Compounds |
Binding Residue |
|
5,7-Dehydroxyflavanone |
Thr259, Phe267, Asn256, Asp258, Tyr271, Arg268, Ile335, Asn331, Ser334, Leu253 |
|
Pinostrombin |
Ile335, Leu253, Tyr271, Phe267, Thr259, Asn256, Ser334, Asn331, Arg268, Asp258 |
|
Chloroquine |
Asp483, Phe485, Glu493, Phe490, Val408, Leu404, Pro405, Val401 |
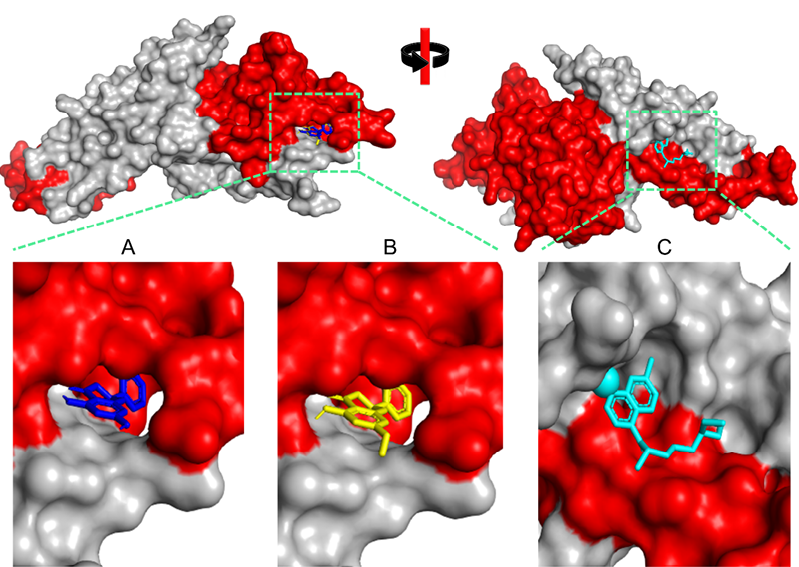
Figure 1. Three-dimensional visualization of Plasmodium vivax Duffy Binding Protein (PvDBP)-ligand complexes. A) 5,7-Dihydroxyflavanonne, B) Pinostrombin, and C) Chloroquine. The red color indicates the DARC binding region.
Two-dimensional visualization pre- and post-MD simulation shows there are alterations of all PvDBP-ligand bonding before and after simulation (Figure 2). Chloroquine lost around 75% of its binding residue towards PvDBP and just retained binding with one DARC residue (Leu404). On the other hand, 5,7-Dyhydroxyflavanone lost its interaction with Phe267 and Arg268 and made new bonds with Asn260. Furthermore, Pinostrobin lost its interaction with two residues (Asn256, Thr259) without making new bonds with other residues.
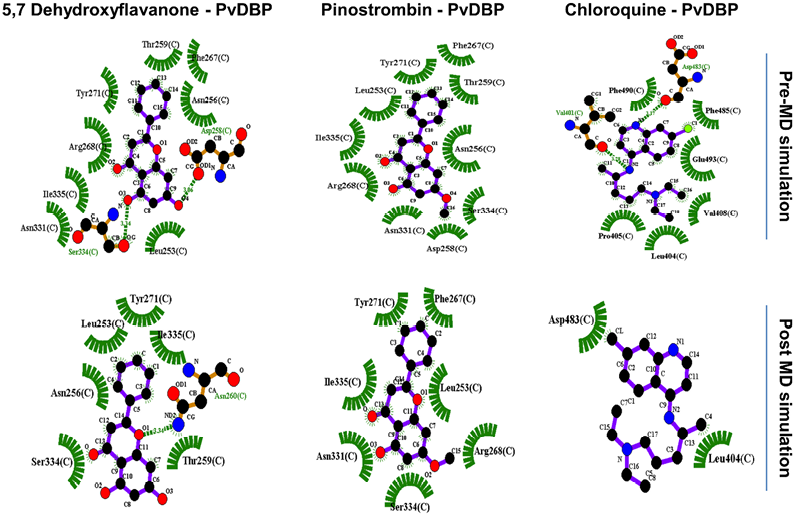
Figure 2. PvDBP-ligand two dimensional visualization before and after 1000 ps molecular dynamic (MD) simulation.
Intramolecular hydrogen bonds do not change significantly over 1000 ps simulation time. PvDBP-5,7-dihydroxyflavanone and PvDBP-chloroquine reach 280 H-bonds in 200 and within 750-800 picoseconds, respectively. On the other hand, the protein-solvent H-bond plot shows that PvDBP-5,7-Dehydroxyflavanone lost its 40 H-bond within 450-500 ps. However, overall, the trends of all three complexes look stable.
The RMSD graph (Figure 3) shows that protein backbones are relatively steady within the 1000 ps simulation after reaching the equilibrium point. The PvDBP – 5,7-dihydroxyflavanone complex has an RMSD value of 1.586 Å, ranging from 0.493 Å to 2.002 Å. Then, the PvDBP-Pinostrobin shows an RMSD value of 1.561 Å, with a maximum score of 1.98 Å and a minimum score of 0.48 Å. Meanwhile, the PvDBP-Chloroquine complex has the highest RMSD score over the 1000 ps simulation 2.302 Å.
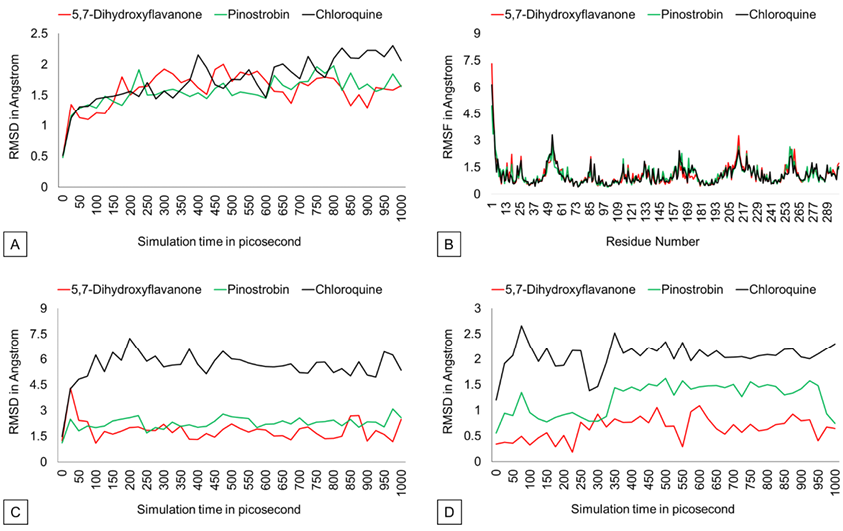
Figure 3. RMSD, RMSF, ligand movement, ligand conformation graph.
The RMSF graph (Figure 3) shows that all three complexes relatively have similar stability over simulation. Highest peaks of 3 complexes were reached by PvDBP-Chloroquine in between 49-61 residues and PvDBP-5,7-dihydroxyflavanone in between 205-217 residues. RMSD Ligand conformation score shows PvDBP-5,7-Dihidroxyflavanone has the lowest value of 0.651Å, whereas PvDBP-chloroquine is the highest. In addition, PvDBP-Chloroquine has the largest value of ligand movement score of 16280.572 Å, which reaches its highest peak of 16712.116Å.
The potential energy value of all protein-ligand complexes is relatively flat over simulation (Figure 4). It indicates that PvDBP-ligand complexes maintain their stability to store potential energy. On the other hand, Protein-ligand binding also does not affect protein expansion. These three complexes reach nearly conformable trends after reaching 400 ps. PvDBP-Chloroquine reached maximum SASA score 16712.116 Å2 at 250 ps. The dynamic cross-correlation maps of three protein-ligand complexes (Figure 5) reveal no significant distinction about the atom’s physical motions. The blue color stands for anti-correlated motions, while yellow represents correlated motions. Thick color stands for high flexibility of protein residues.
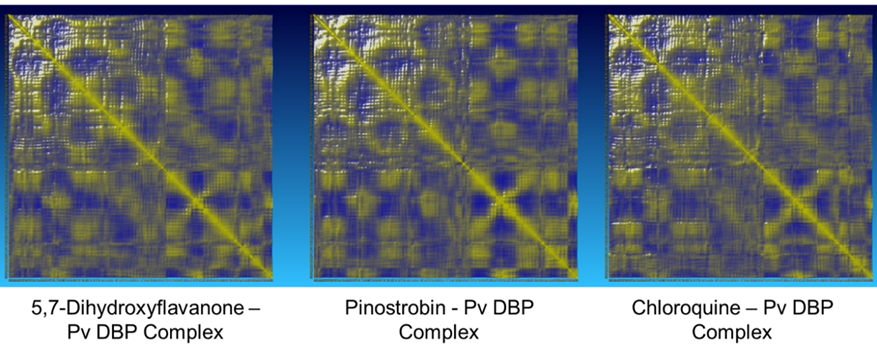
Figure 5. Domain cross-correlation matrix molecular dynamics simulation.
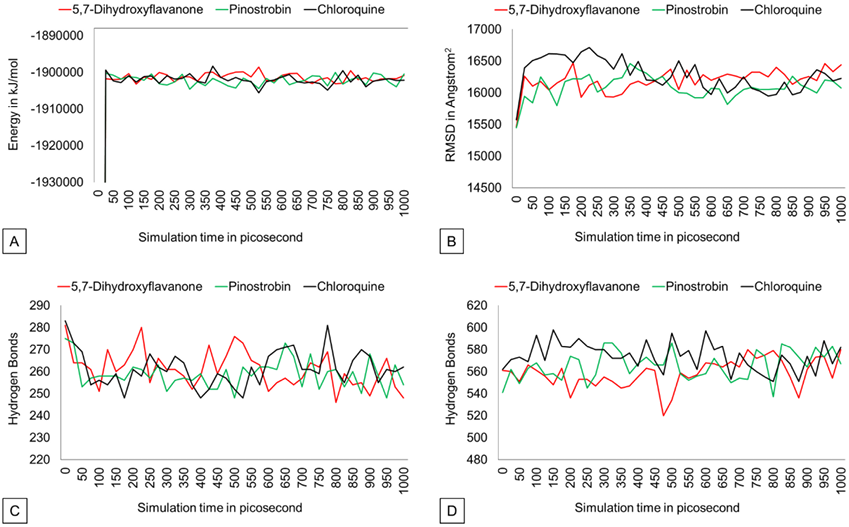
Figure 4. Potential energy, SASA, hydrogen bond (intramolecular), Hydrogen bond solvent (solute) graph.
PvDBP localized in micronemes organelle and presented in Plasmodium vivax surface when merozoite starts the invasion process (Adams et al., 1990; Fraser et al., 1997). PvDBP interacts with DARC through binding with central 170 amino acids in PvDBP region II (RII-PvDBP) (Hans et al., 2005; Singh et al., 2003). PvDBP-DARC molecular interaction is driven by the dimerization process of PvDBP homodimer (Batchelor et al., 2011). Interaction between two molecules of PvDBP resulting in DARC binding groove. In this investigation, all three compounds predicted partially bind into crucial binding residues of DARC.
5,7-dihydroxyflavanone (Pinocembrin) is a flavonoid compound found in the rhizome of Kaempferia rotunda (Atun et al., 2013). The lowest binding energy value towards PvDBP indicates this compound is predicted to have a stronger tendency to incorporate with PvDBP than control. Almost all binding of this compound is located in subdomain 2 of RII-PvDBP. Subdomain 2 of RII-PvDBP is bounded by all DARC-interacted residues (Batchelor et al., 2014). We predicted that the binding stability of 5,7-dihydroxyflavanone was relatively strong with retained bonding with 60% residues and made novel interaction with one DARC binding residue during simulation time. In silico and in vivo studies reveal that pinocembrin and its analog show great antiplasmodial activity (Melaku et al., 2022).
The second compound that may have potential as a drug for malaria is pinostrobin. Pinostrobin is a flavonoid compound that is found in Bosenbergia rotunda (Kanchanapiboon et al., 2020; Morikawa et al., 2008; Tewtrakul, 2005). The binding strength of this compound is not much different from 5,7-dihydroxyflavanone. This compound is predicted to bind to subdomain-1 and subdomain-2 of RII-PvDBP. This compound also binds to Tyr271, whereas this residue plays a role in the PvDBP-DARC dimer interface (Batchelor et al., 2014). Moreover, this pinostrobin-PvDBP interaction is also predicted to disturb the PvDBP-DARC second interface process through bonding with Phe267. Phe267 of RII-PvDBP is required for the DARC binding process and recognized by neutralizing antibodies (Batchelor et al., 2014; Chootong et al., 2010).
Merozoite invasion of red blood cells is required for parasite further development. The parasites digest hemoglobin contained in red blood cells to support their growth (Moore et al., 2006; Pishchany & Skaar, 2012). The PvDBP-compounds interaction in the crucial residues revealed that the DARC-PvDBP association can be disturbed. It affects the internalization process of merozoites in the early blood stage cycle. Inhibition of merozoite internalization possibly controls parasite development extension, throphozoite, schizont, and gametocyte development.
In molecular dynamic analysis, we predicted that 5,7-dihydroxyflavanone-PvDBP and pinostrobin-PvDBP complexes have greater stability than the control. The RMSD value < 2 Å indicates that all three complexes are remarkably stable during simulation time (Castro-Alvarez et al., 2017). Furthermore, protein-ligand binding can lead to protein conformational changes like subdomain and protein backbone movement (Frimurer et al., 2003). Study results reveal that interaction between the two bioactive compounds did not result in a significant conformational alteration of PvDBP. In the RMSF analysis, all three compounds have quite similar effects on binding residue distance. RMSF analysis was used to analyze protein residue mobility during protein-ligand interaction (Zhu et al., 2017). The result indicates that protein-ligand interactions have almost similar binding residue distance fluctuation within the simulation.
All three complexes show stable trends in the SASA value plot. It revealed that ligand-protein binding does not affect the protein’s surface expansion due to the unfolding process (Candotti et al., 2013). Intramolecular hydrogen bonds are required to retain protein structure by maintaining protein secondary structure (Gao et al., 2015; Hubbard & Kamran Haider, 2010). An increase in intramolecular H-bonds indicates that the protein committed a denaturation process by interacting with solvent hydrogen atoms. The number of intramolecular H-bonds of three complexes is relatively stable. This means that the protein-ligand complex did not make significant H-bond number changes. DCCM analysis was used to observe correlative motions of the protein atom during MD simulation (Arnold & Ornstein, 1997; Kasahara et al., 2014). All three complexes convey almost similar flexibility among protein residues.
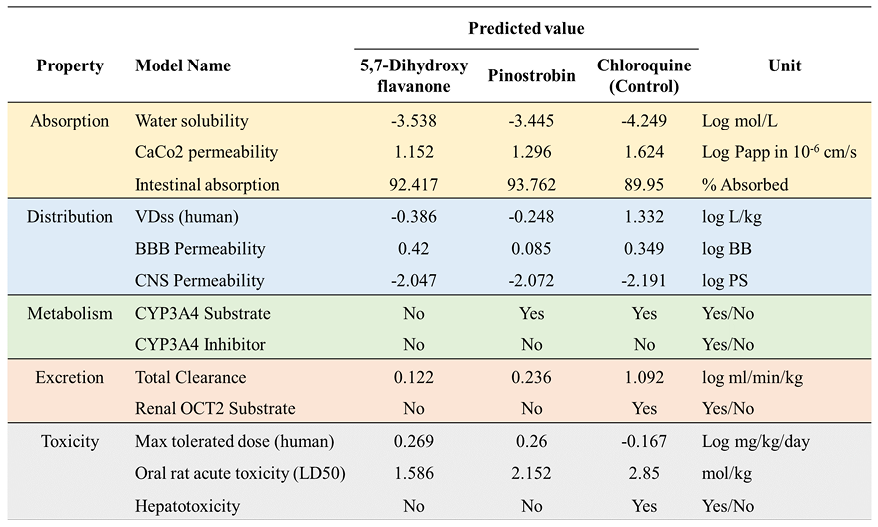
Figure 6. Pharmacokinetics properties prediction of top two compounds and control drug.
Pharmacokinetics properties prediction is important to measure the capabilities of the selected compound to be processed, delivered, and excreted inside the human body. Five criteria were used to determine the pharmacokinetics properties of the selected compound by docking studies (Figure 6). Both top two compounds exhibit good adsorption properties with relatively the same water solubility score, ranging from -3.538 to -4.249. The top two compounds possess higher intestinal absorption compared to chloroquine as a control. CaCo2 permeability indicates that the compounds are easily adsorbed to human colorectal adenocarcinoma cells with the value > 0.90 (Sun et al., 2017). The top two compounds have a low VDss value, while the chloroquine is high (1.332). The findings suggest that the compound is widely distributed in plasma rather than tissue. Chloroquine and 5,7-dihydroxyflavanone were predicted to be readily distributed to the brain due to its logBB score > 0.3. However, among the three compounds, they have a low ability to penetrate the central nervous system. In terms of metabolism properties, the capability of the protein to affect Cytochrome P450 function is considered. This protein acts in the activation and deactivation process of the drugs. There are no compounds that possess the ability as a CYP3A inhibitor. Moreover, only 5,7-dihydroxyflavanone was predicted to influence CYP3A function as a substrate. Chloroquine is predicted to have a higher total clearance score. This parameter is utilized to determine the dosing rate of the compounds to make steady-state concentrations. In toxicity prediction, we found that the two bioactive compounds have less toxicity than chloroquine. These compounds have a higher maximum tolerated dose (human) and lower LD50 scores. Furthermore, 5,7-dihydroxyflavanone and pinostrobin were predicted to not have hepatotoxicity properties. Toxicity prediction assists in reducing experimental costs and time while also addressing ethical issues by considerably lowering animal and clinical testing requirements (Sharma et al., 2023; Cavasotto & Scardino, 2022).
The present study aims to screen numerous Zingiberaceae bioactive compounds as potential inhibitors targetting PvDBP. We found that two compounds (5,7-dihydroxyflavanone and pinostrobin) may prevent PvDBP and DARC interaction due to their strong binding towards DARC binding residues. This research is the first investigation of ligand screening that targets PvDBP as a drug target. Therefore, the findings are important as baseline information for further natural chemistry-based drug development against vivax malaria. Nonetheless, this prediction still requires validation through in vitro and in vivo experiments.
This study was funded by PNBP Universitas Negeri Malang with contract numbers 5.3.619/UN32.14.1/LT/2021 and 5.3.517/UN32.14.1/LT/2021.
The authors thank the Laboratory of Biocomputation, Department of Biology, Brawijaya University, for supporting this study.
The authors declare that there are no conflicts of interest.
All the authors declare that the final version of this paper was read and approved.
The total contribution percentage for the conceptualization, preparation, and correction of this paper was as follows: M.F.H. & W.E.P. 70%; S., A.H., D.W. & M.L 30%.
The data supporting the results of this study will be made available by the corresponding author, [W.E.P.], upon reasonable request.
A Preprint version of this paper was deposited in: https://doi.org/10.5281/zenodo.8307132
Abdou A M, Seddek A S, Abdelmageed N, Badry M O and Nishikawa Y. (2022). Wild Egyptian medicinal plants show in vitro and in vivo cytotoxicity and antimalarial activities. BMC Complementary Medicine and Therapies, 22(1), 130. https://doi.org/10.1186/s12906-022-03566-5
Adams J H, Hudson D E, Torii M, Ward G E, Wellems T E, Aikawa M and Miller L H. (1990). The duffy receptor family of Plasmodium knowlesi is located within the micronemes of invasive malaria merozoites. Cell, 63(1), 141–153. https://doi.org/10.1016/0092-8674(90)90295-P
Ahmed M A, Ameyaw E O, Ackah-Armah F, Acheampong D O, Amoani B, Ampomah P, Adakudugu E A and Adokoh C K. (2022). In vitro and In vivo antimalarial activities of Avicennia africana P. Beauv. (Avicenniaceae) ethanolic leaf extract. Journal of Traditional and Complementary Medicine, 12(4), 391–401. https://doi.org/10.1016/j.jtcme.2021.11.004
Arnold G E and Ornstein R L.(1997). Molecular dynamics study of time-correlated protein domain motions and molecular flexibility: Cytochrome P450BM-3. Biophysical Journal, 73(3), 1147–1159. https://doi.org/10.1016/S0006-3495(97)78147-5
Ashar MSAA, Putra WE, Rifa’i M, Sustiprijatno, Salma WO, Susanto H, Faisal M, Hidayatullah A, Heikal MF, Sholeh M. (2023). Calculating the stability of molecular interface between the ligand-complex and solvent molecule: A study of Averrhoa bilimbi bioactive compounds as anti-diabetic agent. AIP Conference Proceedings 2634, 020023. https://doi.org/10.1063/5.0111215.
Atun S, Arianingrum R, Sulistyowati E and Aznam N. (2013). Isolation and antimutagenic activity of some flavanone compounds from Kaempferia rotunda. International Journal of Chemical and Analytical Science, 4(1): 3–8. https://doi.org/10.1016/j.ijcas.2013.03.004
Batchelor J D, Malpede B M, Omattage N S, DeKoster G T, Henzler-Wildman, K A and Tolia N H. (2014). Red Blood Cell Invasion by Plasmodium vivax: Structural Basis for DBP Engagement of DARC. PLoS Pathogens, 10(1), 908-914. https://doi.org/10.1371/journal.ppat.1003869
Batchelor J D, Zahm J A and Tolia N H. (2011). Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nature Structural & Molecular Biology, 18(8), 908–914. https://doi.org/10.1038/nsmb.2088
Battle K E and Baird J K. (2021). The global burden of Plasmodium vivax malaria is obscure and insidious. PLOS Medicine, 18(10), 1-18. https://doi.org/10.1371/journal.pmed.1003799
Candotti M, Pérez A, Ferrer-Costa C, Rueda M, Meyer T, Gelpí J L and Orozco M. (2013). Exploring Early Stages of the Chemical Unfolding of Proteins at the Proteome Scale. PLoS Computational Biology, 9(12), e1003393. https://doi.org/10.1371/journal.pcbi.1003393
Castro-Alvarez A, Costa A and Vilarrasa J. (2017). The Performance of Several Docking Programs at Reproducing Protein–Macrolide-Like Crystal Structures. Molecules, 22(1), 136. https://doi.org/10.3390/molecules22010136
Cavasotto CN, Scardino V. (2022). Machine learning toxicity prediction: latest advances by toxicity end point. ACS Omega, 7(51), 47536-47546. https://doi.org/10.1021/acsomega.2c05693
Chootong P, Ntumngia F B, VanBuskirk K M, Xainli J, Cole-Tobian J L, Campbell C O, Fraser T S, King C L and Adams J H. (2010). Mapping Epitopes of the Plasmodium vivax Duffy Binding Protein with Naturally Acquired Inhibitory Antibodies. Infection and Immunity, 78(3), 1089–1095. https://doi.org/10.1128/IAI.01036-09
Chu C S and White N J. (2021). The prevention and treatment of Plasmodium vivax malaria. PLOS Medicine, 18(4), 1-21. https://doi.org/10.1371/journal.pmed.1003561
Dini S, Douglas N M, Poespoprodjo J R, Kenangalem E, Sugiarto P, Plumb I D, Price R N and Simpson J A. (2020). The risk of morbidity and mortality following recurrent malaria in Papua, Indonesia: A retrospective cohort study. BMC Medicine, 18(1), 1-12. https://doi.org/10.1186/s12916-020-1497-0
Ferreira M U, Nobrega de Sousa T, Rangel G W, Johansen I C, Corder R M, Ladeia-Andrade S and Gil J P. (2021). Monitoring Plasmodium vivax resistance to antimalarials: Persisting challenges and future directions. International Journal for Parasitology: Drugs and Drug Resistance, 15, 9–24. https://doi.org/10.1016/j.ijpddr.2020.12.001
Fraser T, Michon P, Barnwell J W, Noe A R, Al-Yaman F, Kaslow D C and Adams J H. (1997). Expression and serologic activity of a soluble recombinant Plasmodium vivax Duffy binding protein. Infection and Immunity, 65(7), 2772–2777. https://doi.org/10.1128/iai.65.7.2772-2777.1997
Frimurer T M, Peters G H, Iversen L F, Andersen H S, Møller N P H and Olsen O H. (2003). Ligand-Induced Conformational Changes: Improved Predictions of Ligand Binding Conformations and Affinities. Biophysical Journal, 84(4), 2273–2281. https://doi.org/10.1016/S0006-3495(03)75033-4
Gao Y, Mei Y and Zhang J Z H. (2015). Treatment of Hydrogen Bonds in Protein Simulations. In J. Liu (Ed.), Advanced Materials for Renewable Hydrogen Production, Storage and Utilization, 25, 121-136. https://doi.org/10.5772/61049
Getachew S, Thriemer K, Auburn S, Abera A, Gadisa E, Aseffa A, Price R N and Petros B. (2015). Chloroquine efficacy for Plasmodium vivax malaria treatment in southern Ethiopia. Malaria Journal, 14(1), 1-8. https://doi.org/10.1186/s12936-015-1041-4
Han J H, Lee S K, Wang B, Muh F, Nyunt M H, Na S, Ha K S, Hong S H, Park W S, Sattabongkot J, Tsuboi T and Han E T. (2016). Identification of a reticulocyte-specific binding domain of Plasmodium vivax reticulocyte-binding protein 1 that is homologous to the PfRh4 erythrocyte-binding domain. Scientific Reports, 6(1), 1-12. https://doi.org/10.1038/srep26993
Hans D, Pattnaik P, Bhattacharyya A, Shakri A R, Yazdani S S, Sharma M, Choe H, Farzan, M and Chitnis C E. (2005). Mapping binding residues in the Plasmodium vivax domain that binds Duffy antigen during red cell invasion: Binding residues of P. vivax Duffy binding protein. Molecular Microbiology, 55(5), 1423–1434. https://doi.org/10.1111/j.1365-2958.2005.04484.x
Heikal MF, Putra WE, Sustiprijatno, Rifa’i M, Hidayatullah A, Ningsih FN, Widiastuti D, Shuib AS, Zulfiani BF, Hanasepti AF. (2023). In silico screening and molecular dynamics simulation of potential anti-malarial agents from Zingiberaceae as potential Plasmodium falciparum lactate dehydrogenase (PfLDH) enzyme inhibitors. Tropical Life Sciences Research, 34(2), 1-20. https://doi.org/10.21315/tlsr2023.34.2.1
Hidayatullah A, Putra WE, Rifa’i M, Sustiprijatno, Widiastuti D, Heikal MF, Susanto H, Salma WO. (2022). Molecular docking and dynamics simulation studies to predict multiple medicinal plants’ bioactive compounds interaction and its behavior on the surface of DENV-2 E protein. Karbala International Journal of Modern Science, 8(3), 531-542. https://doi.org/10.33640/2405-609X.3237
Hidayatullah A, Putra WE, Salma WO, Muchtaromah B, Permatasari GW, Susanto H, Widiastuti D, Kismurtono M. (2021). Discovery of drug candidate from various natural products as potential novel dengue virus nonstructural protein 5 (NS5) inhibitor. Chiang Mai University Journal of Natural Sciences, 20(1), 1-17. https://doi.org/10.12982/CMUJNS.2021.018
Hidayatullah A, Putra WE, Sustiprijatno, Permatasari GW, Salma WO, Widiastuti D, Susanto H, Muchtaromah B, Sari DRT, Ningsih FN, Heikal MF, Yusuf AMR, Arizona AS. (2021). In silico targeting DENV2’s prefusion envelope protein by several natural products’ bioactive compounds. Chiang Mai University Journal of Natural Sciences, 20(4), 1-20.
Hidayatullah A, Putra WE, Sustiprijatno, Widiastuti D, Salma WO, Heikal MF. (2023). Molecular docking and molecular dynamics simulation-based identification of natural inhibitors against druggable human papilloma virus type 16 target. Trends in Sciences, 20(4), 1-12. https://doi.org/10.48048/tis.2023.4891
Howes R E, Battle K E, Mendis K N, Smith D L, Cibulskis R E, Baird J K and Hay S I. (2016). Global Epidemiology of Plasmodium vivax. The American Journal of Tropical Medicine and Hygiene, 95(6), 15–34. https://doi.org/10.4269/ajtmh.16-0141
Htun M W, Mon N C N, Aye K M, Hlaing C M, Kyaw M P, Handayuni I, Trimarsanto H, Bustos D, Ringwald P, Price R N, Auburn S and Thriemer K. (2017). Chloroquine efficacy for Plasmodium vivax in Myanmar in populations with high genetic diversity and moderate parasite gene flow. Malaria Journal, 6(1), 281. https://doi.org/10.1186/s12936-017-1912-y
Hubbard R E and Kamran Haider M. (2010). Hydrogen Bonds in Proteins: Role and Strength. In John Wiley & Sons, Ltd (Ed.) ELS (1st ed.). Wiley. https://doi.org/10.1002/9780470015902.a0003011.pub2
Kanchanapiboon J, Kongsa U, Pattamadilok D, Kamponchaidet S, Wachisunthon D, Poonsatha S and Tuntoaw S. (2020). Boesenbergia rotunda extract inhibits Candida albicans biofilm formation by pinostrobin and pinocembrin. Journal of Ethnopharmacology, 261, 1-9. https://doi.org/10.1016/j.jep.2020.113193
Kasahara K, Fukuda I and Nakamura H. (2014). A Novel Approach of Dynamic Cross Correlation Analysis on Molecular Dynamics Simulations and Its Application to Ets1 Dimer–DNA Complex. PLoS ONE, 9(11), 1-13. https://doi.org/10.1371/journal.pone.0112419
Korsinczky M, Fischer K, Chen N, Baker J, Rieckmann K and Cheng Q. (2004). Sulfadoxine Resistance in Plasmodium vivax Is Associated with a Specific Amino Acid in Dihydropteroate Synthase at the Putative Sulfadoxine-Binding Site. Antimicrobial Agents and Chemotherapy, 48(6), 2214–2222. https://doi.org/10.1128/AAC.48.6.2214-2222.2004
Lipinski C A. (2004). Lead- and drug-like compounds: The rule-of-five revolution. Drug Discovery Today: Technologies, 1(4), 337–341. https://doi.org/10.1016/j.ddtec.2004.11.007
Markus M B. (2011). Malaria: Origin of the Term “Hypnozoite.” Journal of the History of Biology 44(4), 781–786. https://doi.org/10.1007/s10739-010-9239-3
Maslikah SI, Putra WE. (2024). Molecular dynamics simulation of various bioactive compounds of red betel (Piper crocatum) as anti-inflammatory drug-like candidates in rheumatoid arthritis treatment. Trends in Sciences, 21(3), 1-8. https://doi.org/10.48048/tis.2024.7334
Melaku Y, Solomon M, Eswaramoorthy R, Beifuss U, Ondrus V and Mekonnen Y. (2022). Synthesis, antiplasmodial activity and in silico molecular docking study of pinocembrin and its analogs. BMC Chemistry, 16(1), 36. https://doi.org/10.1186/s13065-022-00831-z
Moore L R, Fujioka H, Williams P S, Chalmers J J, Grimberg B, Zimmerman P A and Zborowski M. (2006). Hemoglobin degradation in malaria-infected erythrocytes determined from live cell magnetophoresis. The FASEB Journal, 20(6), 747–749. https://doi.org/10.1096/fj.05-5122fje
Morikawa T, Funakoshi K, Ninomiya K, Yasuda D, Miyagawa K, Matsuda H and Yoshikawa M. (2008). Medicinal Foodstuffs. XXXIV.1) Structures of New Prenylchalcones and Prenylflavanones with TNF-α and Aminopeptidase N Inhibitory Activities from Boesenbergia rotunda. Chemical and Pharmaceutical bulletin, 56(7), 956-962. https://doi.org/10.1248/cpb.56.956
Okokon J E, Mobley R, Edem U A, Bassey A I, Fadayomi I, Drijfhout F, Horrocks P and Li W W. (2022). In vitro and in vivo antimalarial activity and chemical profiling of sugarcane leaves. Scientific Reports, 12(1), 1-13. https://doi.org/10.1038/s41598-022-14391-8
Ould Ahmedou Salem M S, Mohamed Lemine Y O, Deida J M, Lemrabott M A O, Ouldabdallahi M, Ba M, dit D, Boukhary A O M S, Khairy M L O, Abdel Aziz M B, Ringwald P, Basco L K, Niang S D and Lebatt S M. (2015). Efficacy of chloroquine for the treatment of Plasmodium vivax in the Saharan zone in Mauritania. Malaria Journal, 14(1), 1-5. https://doi.org/10.1186/s12936-015-0563-0
Pishchany G and Skaar E P. (2012). Taste for Blood: Hemoglobin as a Nutrient Source for Pathogens. PLoS Pathogens, 8(3), 1-14. https://doi.org/10.1371/journal.ppat.1002535
Putra WE, Rifa’i M. (2020). Evaluating the molecular interaction of sambucus plant bioactive compounds toward TNF-R1 and TRAIL-R1/R2 as possible anti-cancer therapy based on traditional medicine: The bioinformatics study. Scientific Study & Research - Chemistry & Chemical Engineering, Biotechnology, Food Industry, 21(3), 357-365.
Putra WE, Salma WO, Rifa’i M. (2019). Anti-inflammatory activity of sambucus plant bioactive compounds against TNF-α and TRAIL as solution to overcome inflammation associated diseases: The insight from bioinformatics study. Natural Product Sciences, 25(3), 215-221. https://doi.org/10.20307/nps.2019.25.3.215
Putra WE, Salma WO, Widiastuti D, Kismurtono M. (2020). In silico screening of peroxisome proliferator activated receptor gamma (PPARG)-agonist from Eugenia jambolana bioactive compounds as potential anti-diabetic agent. Malaysian Journal of Biochemistry & Molecular Biology, 23(2), 142-146.
Putra WE, Sustiprijatno, Hidayatullah A, Heikal MF, Widiastuti D, Isnanto H. (2023). Analysis of three non-structural proteins, Nsp1, Nsp2, and Nsp10 of Sars-Cov-2 as pivotal target proteins for computational drug screening. Journal of Microbiology, Biotechnology and Food Sciences, 12(5), 1-6. https://doi.org/10.55251/jmbfs.9586
Putra WE, Waffareta E, Ardiana O, Januarisasi ID, Soewondo A, Rifa’i M. (2017). Dexamethasone-administrated BALB/c mouse promotes proinflammatory cytokine expression and reduces CD4+CD25+ regulatory T cells population. Bioscience Research, 2017. 14(2), 201-213.
Putra WE. (2018). In silico study demonstrates multiple bioactive compounds of sambucus plant promote death cell signaling pathway via Fas receptor. FUW Trends in Science & Technology Journal, 3(2), 682-685.
Ratcliff A, Siswantoro H, Kenangalem E, Maristela R, Wuwung R, Laihad F, Ebsworth E, Anstey N, Tjitra E and Price R. (2007). Two fixed-dose artemisinin combinations for drug-resistant falciparum and vivax malaria in Papua, Indonesia: An open-label randomised comparison. The Lancet, 369(9563), 757–765. https://doi.org/10.1016/S0140-6736(07)60160-3
Sari AN, Putra WE, Rifa’i M, Sustiprijatno, Susanto H, Salma WO, Faisal M, Hidayatullah A, Heikal MF, Firdaus SRA. (2023). Profiling the coulomb energy of chimanine D and desulphosinigrin as potential anti-diabetic drug against alpha-glucosidase. AIP Conference Proceedings 2634, 020012. https://doi.org/10.1063/5.0111214
Sharma B, Chenthamarakshan V, Dhurandhar A, Pereira S, Hendler JA, Dordick JS, Das P. (2023). Accurate clinical toxicity prediction using multi-task deep neural nets and contrastive molecular explanations. Scientific Reports, 13(1), 1-16. https://doi.org/10.1038/s41598-023-31169-8
Singh S K, Singh A P, Pandey S, Yazdani S S, Chitnis C E and Sharma A. (2003). Definition of structural elements in Plasmodium vivax and P. knowlesi Duffy-binding domains necessary for erythrocyte invasion. Biochemical Journal, 374(1), 193–198. https://doi.org/10.1042/bj20030622
Sun H, Nguyen K, Kerns E, Yan Z, Yu KR, Shah P, Jadhav A, Xu X. (2017). Highly predictive and interpretable models for PAMPA permeability. Bioorganic & Medicinal Chemistry, 25(3), 1266-1276. https://doi.org/10.1016/j.bmc.2016.12.049
Sutanto I, Endawati D, Ling L H, Laihad F, Setiabudy R and Baird J K. (2010). Evaluation of chloroquine therapy for vivax and falciparum malaria in southern Sumatra, western Indonesia. Malaria Journal, 9(1), 52. https://doi.org/10.1186/1475-2875-9-52
Tewtrakul S, Subhadhirasakul S and Kummee S. (2005). HIV-1 protease inhibitory effects of medicinal plants used as self medication by AIDS patients. Songklanakarin Journal of Science and Technology, 25(2), 239-243
Widiastuti D, Warnasih S, Sinaga SE, Pujiyawati E, Supriatno, Putra WE. (2023). Identification of active compounds from Averrhoa bilimbi L. (Belimbing Wuluh) flower using LC-MS and antidiabetic activity test using in vitro and in silico approaches. Trends in Sciences, 20(8), 1-9. https://doi.org/10.48048/tis.2023.6761
World Health Organization. (2020). World Malaria Report: 20 years of global progress and challenges. World Health Organization.
Zhu J, Lv Y, Han X, Xu D and Han W. (2017). Understanding the differences of the ligand binding/unbinding pathways between phosphorylated and non-phosphorylated ARH1 using molecular dynamics simulations. Scientific Reports, 7(1), 1-14. https://doi.org/10.1038/s41598-017-12031-0
1* Corresponding author
Muhammad Fikri Heikal,
 muhammadfikriheikal@gmail.com,
muhammadfikriheikal@gmail.com,  http://orcid.org/0000-0002-4672-8107
http://orcid.org/0000-0002-4672-8107Wira Eka Putra,
 wira.putra.fmipa@um.ac.id,
wira.putra.fmipa@um.ac.id,  http://orcid.org/0000-0003-4831-3869
http://orcid.org/0000-0003-4831-3869Sustiprijatno,
 susti11.prijatno@gmail.com,
susti11.prijatno@gmail.com,  http://orcid.org/0000-0001-8623-8471
http://orcid.org/0000-0001-8623-8471Arief Hidayatullah,
 arief.hidayatullah37@gmail.com,
arief.hidayatullah37@gmail.com,  http://orcid.org/0000-0003-1929-3635
http://orcid.org/0000-0003-1929-3635Diana Widiastuti,
 dianawidi25@unpak.ac.id,
dianawidi25@unpak.ac.id,  http://orcid.org/0000-0001-6844-3906
http://orcid.org/0000-0001-6844-3906Mardiana Lelitawati,
 mardiana.lelitawati.fmipa@um.ac.id,
mardiana.lelitawati.fmipa@um.ac.id,  https://orcid.org/0000-0002-2249-1192
https://orcid.org/0000-0002-2249-1192Department of Biology, Faculty of Mathematics and Natural Sciences, Universitas Negeri Malang, East Java, Indonesia.
2 Biotechnology Study Program, Department of Applied Sciences, Faculty of Mathematics and Natural Sciences, Universitas Negeri Malang, East Java, Indonesia.
3 Research Center for Applied Botany, National Research and Innovation Agency West Java, Indonesia.
4 Health Governance Initiative, United Nations Development Programme Indonesia, Eijkman-RSCM Building, Jakarta, Indonesia
5 Department of Chemistry, Faculty of Mathematics and Natural Science, Universitas Pakuan, West Java, Indonesia.
A Molecular Docking and Dynamics Simulation Study on Prevention of Merozoite Red Blood Cell Invasion by Targeting Plasmodium vivax Duffy Binding Protein with Zingiberaceae Bioactive Compounds (Muhammad Fikri Heikal • Wira Eka Putra • Sustiprijatno • Arief Hidayatullah • Diana Widiastuti • Mardiana Lelitawati) Uniciencia is protected by Attribution-NonCommercial-NoDerivs 3.0 Unported (CC BY-NC-ND 3.0)
URL: www.revistas.una.ac.cr/uniciencia
Correo electrónico: revistauniciencia@una.cr


